周志华西瓜书3.4python代码
选择数据集
乳腺癌
编程参考资料:
针对Breast-Cancer(乳腺癌)数据集
数据集划分方法
他人答案
自己写代码主要还是熟悉怎么调包,练练手先。写了乳腺癌的代码,跑了发现两个问题:
1.10-fold CV比不过别人[普通的划分方法](https://www.bbsmax.com/A/QW5YW18Mzm/)。
2.LOO估计出来的正确率为0

以下是乳腺癌的原代码,别急着用
#DATASET#1:乳腺癌
import numpy as np
import pandas as pd
from sklearn.metrics import classification_report
#以下代码参考https://www.bbsmax.com/A/QW5YW18Mzm/
# 创建每列名字
columnNames = [
'Sample code number',
'Clump Thickness',
'Uniformity of Cell Size',
'Uniformity of Cell Shape',
'Marginal Adhesion',
'Single Epithelial Cell Size',
'Bare Nuclei',
'Bland Chromatin',
'Normal Nucleoli',
'Mitoses',
'Class'
]
data = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',names = columnNames)#如果是LOO还要补出参数: delim_whitespace=True
# 清洗空缺数据
data = data.replace(to_replace = "?", value = np.nan)#将丢失数据代替成 "?"
data = data.dropna(how = 'any')# 然后丢掉他们
X = data.iloc[:,0:10]
Y = data.iloc[:,10]
##以下代码参考https://blog.csdn.net/Snoopy_Yuan/article/details/64131129
#对率回归
from sklearn.linear_model import LogisticRegression
#metrics是评估模块,例如准确率等
from sklearn import metrics
from sklearn.model_selection import cross_val_predict
log_model=LogisticRegression()
'''
#10fold-CV,cross_val_predict返回的是estimator的分类结果,用于和实际数据比较
Y_pred = cross_val_predict(log_model,X,Y,cv=10)
print("iris with 10folds, precision is:",metrics.accuracy_score(Y,Y_pred))
'''
'''
#--------------------------------方法分割线------------------------------------------
#LOOCV
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
accuracy = 0#因为只有一个样本,所以默认为0
#split是leave-one-out模型的方法,把数据分隔为train和test数组
for train,test in loo.split(X):
log_model.fit(X[train],Y[train]) #fit模型
Y_p=log_model.predict(X[test])
if Y_p==Y[test]:
accuracy+=1
print("iris with LeaveOneOut, precision is:",accuracy/np.shape(X)[0]) #shape(x)是数组维度,shape(x)[0]相当于数组的行数也就是样本数
'''
现在检查哪里出问题了ε=(´ο`*)))
因为代码都是搬运的,大佬们实验么得问题,所以分块核对:
1.针对block#1复核他人代码,不同之处有三:
X = data.iloc[:,0:10]
Y = data.iloc[:,10]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, # features
Y, # labels
test_size = 0.1,
random_state = 33
)
输出:
Accuracy of the LogesticRegression: 0.5217391304347826
OK,方法一错在哪里找出来了。现在修改代码为:
Y_pred = cross_val_predict(log_model,
data[ columnNames[1:10] ], # features
data[ columnNames[10] ], # labels
cv=10)
虽然报了一堆警告,但是输出是
breast-cancer-wisconsin with 10folds, precision is: 0.9604685212298683
好歹结果差不多,报了那么多警告纯粹是因为10折跑了10次。这是可以跑的代码:
# -*- coding: utf-8 -*-
"""
Created on Fri Feb 14 17:31:35 2020
@author: 29033
"""
#DATASET#1:乳腺癌
import numpy as np
import pandas as pd
from sklearn.metrics import classification_report
# 创建每列名字
columnNames = [
'Sample code number',
'Clump Thickness',
'Uniformity of Cell Size',
'Uniformity of Cell Shape',
'Marginal Adhesion',
'Single Epithelial Cell Size',
'Bare Nuclei',
'Bland Chromatin',
'Normal Nucleoli',
'Mitoses',
'Class'
]
data = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',names = columnNames)
# 数据处理
data = data.replace(to_replace = "?", value = np.nan)#将丢失数据代替成 "?"
data = data.dropna(how = 'any')# 然后丢掉他们
X = data[ columnNames[1:10] ]# features
Y = data[ columnNames[10] ]# labels
#对率回归
from sklearn.linear_model import LogisticRegression
#metrics是评估模块,例如准确率等
from sklearn import metrics
from sklearn.model_selection import cross_val_predict
log_model=LogisticRegression()
#10折交叉验证
Y_pred = cross_val_predict(log_model,X,Y,cv=10)
print("breast-cancer-wisconsin with 10folds, precision is:",metrics.accuracy_score(Y,Y_pred))
现在针对问题2。修改后跑了,得出正确率为 0.9633967789165446【就是警告有些多】:
#--------------------------------方法分割线------------------------------------------
#留一法
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
accuracy = 0#因为只有一个样本,所以默认为0
#split是leave-one-out模型的方法,把数据分隔为train和test数组
for train,test in loo.split(X):#loo.split(X)的类型是
#train每回有682个,出现683次,类型是
log_model.fit(X.iloc[train], Y.iloc[train]) # fitting
Y_p = log_model.predict(X.iloc[test])
if (Y_p == Y.iloc[test]).any() :
accuracy += 1
print("For the LOOCV, precision is:",accuracy/np.shape(X)[0]) #shape(x)是数组维度,shape(x)[0]相当于数组的行数也就是样本数
回顾一下,发现是切片出错。X1是之前错误的切片方式,比较一下有:
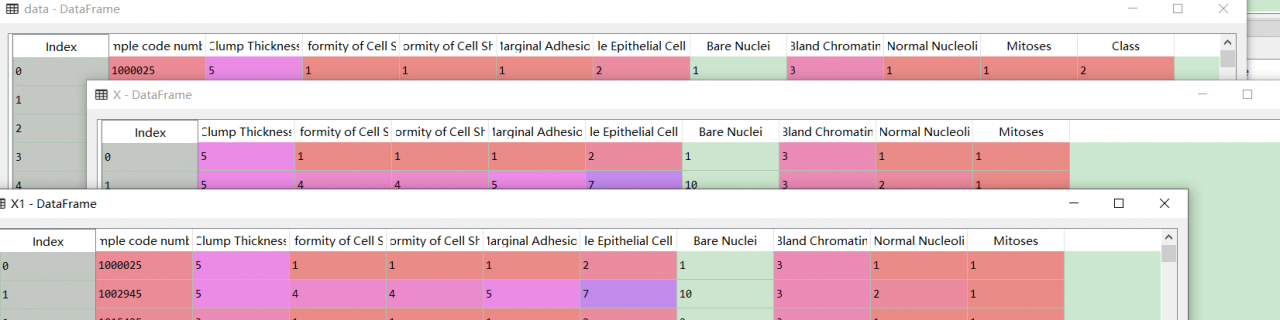 应该改成是
应该改成是
X2 = data.iloc[:,1:10]
Y2 = data.iloc[:,10]
核对后没毛病(๑•̀ㅂ•́)و✧原来的代码改一改也可以跑。LOOCV核对,因为特征矩阵和标签是对应的,所以划分后仍然对称,没有问题
被生活毒打一顿后选择数据集iris╮(╯▽╰)╭虽然有现成的,但建议自己默写代码增强熟练度。
作者:shianlin2084