随机森林实现及调参的R与Python对比——以泰坦尼克幸存者数据为例
随机森林实现及调参一、R语言方法一、手动调参方法二、网格调参二、python
注:本博客数据仍采用决策树调参的泰坦尼克号数据,前奏(数据预处理)请参考☞
决策树R&Python调参对比☜ 一、R语言 方法一、手动调参
作者:whether-or-not
注:本博客数据仍采用决策树调参的泰坦尼克号数据,前奏(数据预处理)请参考☞
决策树R&Python调参对比☜ 一、R语言 方法一、手动调参
PS.仅使用常规包:randomForest和循环编写。
1-建模
set.seed(6)
rf <- randomForest(Survived~.,data=train,ntree=100)
y_pred <- predict(rf,test)
A <- as.matrix(table(y_pred,test$Survived))
acc <- sum(diag(A))/sum(A);acc
未做任何处理时模型精度达到0.8345865。(甚至超过决策树python调参后的结果)
2-1调参——特征数
err <- as.numeric()
for(i in 1:(ncol(train)-1)){
set.seed(6)
mtry_n <- randomForest(Survived~.,data=train,mtry=i)
err <- append(err,mean(mtry_n$err.rate))
}
print(err)
mtry <- which.min(err);mtry
2-2调参——树的个数
set.seed(6)
ntree_fit <- randomForest(Survived~.,data=train,mtry=mtry,
ntree=400)
plot(ntree_fit)
set.seed(0219)
fold <- createFolds(y = data$Survived, k=10)
right <- as.numeric()
for (i in 40:100){
accuracy <- as.numeric()
for(j in 1:10){
fold_test <- data[fold[[j]],]
fold_train <- data[-fold[[j]],]
set.seed(1234)
fold_fit <- randomForest(Survived~.,data=fold_train,mtry=mtry,
ntree=i)
fold_pred <- predict(fold_fit,fold_test)
confumat <- as.matrix(table(fold_pred,fold_test$Survived))
acc <- sum(diag(confumat))/sum(confumat)
accuracy = append(accuracy,acc)
}
right <- append(right,mean(accuracy))
}
print(max(right))
print(which.max(right)+40)
本段结合交叉验证的随机森林树个数的调参为博主自行编写,若有问题请私信讨论,转摘请注明出处,谢谢!
3-最优模型预测
set.seed(6)
rf_best <- randomForest(Survived~.,data=train,mtry=3,ntree=58)
pred <- predict(rf_best,test)
A <- as.matrix(table(pred,test$Survived))
acc <- sum(diag(A))/sum(A);acc
特征数和基分类器的个数调整后模型精度达到0.8609023!!!比未调整结果提高约2.5%!!!
方法二、网格调参PS.使用强大的caret包和trainControl、tunegrid函数。
但随机森林网格调参只有一个参数mtry,且为随机调整.
library(caret)
metric = "Accuracy"
control <- trainControl(method = "repeatedcv", number = 10, repeats = 10)
set.seed(6)
rf_carte <- train(Survived~.,data=train,method = "rf",
metric = "Accuracy", trControl = control,
search = "random")
modelLookup(model = "rf")
rf_carte
y <- predict(rf_carte,test
#,type = "prob"
)
A <- as.matrix(table(y,test$Survived))
acc <- sum(diag(A))/sum(A);acc # 0.8345865
特征数减少,模型泛化效果变差,此时模型误差上升,出现过拟合。
PS.若小伙伴有关于随机森林网格调参更好的办法或更多关于caret包的资料,欢迎一起学习讨论!
0-导入所需库(决策树调参已导入的不再导入)
from sklearn.ensemble import RandomForestClassifier
1-建模
rf = RandomForestClassifier(n_estimators=100,random_state=19)
rf.cv = cross_val_score(rf,x,y,cv=10).mean()
print(rf.cv)
rf0 = rf.fit(xtrain,ytrain)
score = rf0.score(xtest,ytest)
print(score)
2-调参
2-1 n_estimators
best_ntree = []
for i in range(1,201,10):
rf = RandomForestClassifier(n_estimators=i,
n_jobs=-1,
random_state = 19)
score = cross_val_score(rf,x,y,cv=10).mean()
best_ntree.append(score)
print(max(best_ntree),np.argmax(best_ntree)*10)
plt.figure()
plt.plot(range(1,201,10),best_ntree)
plt.show()
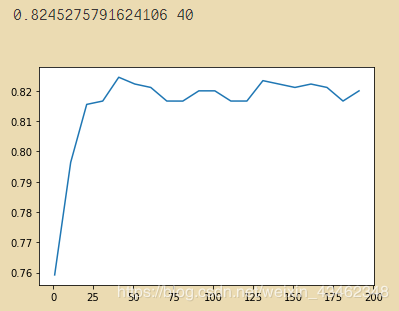
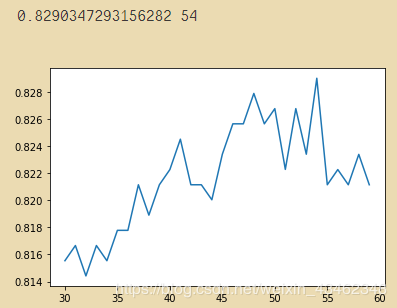
# 缩短区间查看:
ntree = []
for i in range(30,60):
rf = RandomForestClassifier(n_estimators=i
,random_state=19
,n_jobs=-1)
score = cross_val_score(rf,x,y,cv=10).mean()
ntree.append(score)
print(max(ntree),np.argmax(ntree)+30) # ntree = 55
plt.plot(range(30,60),ntree)
plt.show()
6-调参(2)max_depth
from sklearn.model_selection import GridSearchCV
param_grid = {'max_depth':[*range(1, 9)]} # 设置参数
rf = RandomForestClassifier(n_estimators=55
,random_state=19
)
GS = GridSearchCV(rf,param_grid,cv=10)
GS.fit(x,y)
GS.best_score_ # 0.8335208098987626
GS.best_params_ # max_depth=8
6-调参(3)max_features
param_grid = {'max_features':[*range(1,4)]}
rf = RandomForestClassifier(n_estimators=55
,random_state=19
,max_depth=8
)
GS = GridSearchCV(rf,param_grid,cv=10)
GS.fit(x,y)
GS.best_score_ # 0.8335208098987626
GS.best_params_ # max_features=3
6-调参(4)min_samples_leaf,min_samples_split
param_grid = {'min_samples_leaf':[*range(1,11)],'min_samples_split':[*range(2,22)]}
rf = RandomForestClassifier(n_estimators=55
,random_state=19
,max_depth=8
,max_features=3
)
GS = GridSearchCV(rf,param_grid,cv=10)
GS.fit(x,y)
GS.best_score_ # 0.8368953880764904
GS.best_params_ # min_samples_leaf=1,min_samples_split=4
本人也试了所有参数整体调参,但费时很长,有兴趣的小伙伴可以试试。
# 整体调参
param_grid = {'max_depth':[*range(1,9)],'max_features':[*range(1,9)]
,'min_samples_leaf':[*range(1,11)],'min_samples_split':[*range(2,22)]}
rf = RandomForestClassifier(n_estimators=55
,random_state=19
)
GS = GridSearchCV(rf,param_grid,cv=10)
GS.fit(x,y)
GS.best_score_ # 0.8402699662542182
综上,此时单独调参的训练结果得到的最优模型交叉验证的准确率约为0.8369;
整体调参可达到约0.8403。
R网格调参结果约为:0.8346;
R手动调参结果约为:0.8609,R手动调参结果最优,而网格调参可调整的参数有限仅能达到0.8346低于python网格调参,python的sklearn库调参可操作空间大。
作者:whether-or-not